De Novo Design Of Protein Structure And Function With Rfdiffusion
De Novo Design Of Protein Structure And Function With Rfdiffusion - Web we demonstrate the power and generality of the method, called rosettafold diffusion (rfdiffusion), by experimentally characterizing the structures. Web ligadvisor is a flexible and intuitive tool that supports researchers in the de novo discovery and design of drugs 17. Web protein design is founded on inverting this process: Web a team led by scientists from the baker lab has created a powerful new way of designing proteins that combines structure prediction networks and generative. Web rfdiffusion is a software that enables the design of novel proteins with desired structure and function. Web we demonstrate the power and generality of the method, called rosettafold diffusion (rfdiffusion), by experimentally characterizing the structures. Sbgrid webinars are hosted with partial support from the nih r25 continuing education for structural. Web this perspective discusses the concepts and approaches of de novo protein design, emerging challenges in designing structure and function, and the frontiers that. Web rfdiffusion is a method that uses diffusion models to generate protein backbones with various functions and symmetries. The de novo assembly of the wt transcriptome was. Web notably, the model generalizes strongly, accurately predicting mutation effects for proteins with low sequence homology and de novo designed proteins — areas. Web a team led by scientists from the baker lab has created a powerful new way of designing proteins that combines structure prediction networks and generative. Sbgrid webinars are hosted with partial support from the nih r25. Web de novo design of protein structure and function with rfdiffusion jason yim · brian l trippe abstract: Web de novo protein design seeks to generate proteins with specified structural and/or functional properties, for example, making a binding interaction with a given. Web protein design is founded on inverting this process: Web a team led by scientists from the baker. Web we demonstrate the power and generality of the method, called rosettafold diffusion (rfdiffusion), by experimentally characterizing the structures. The de novo assembly of the wt transcriptome was. Web the new algorithm builds on the authors’ previous rosettafold method by adding a diffusion model that enables de novo design of protein backbones with. While this approach has successfully generated new. Web instead, beta barrels and other protein folds have been designed guided by 2d structural blueprints; While this approach has successfully generated new fluorescent. Web rfdiffusion is a method that uses diffusion models to generate protein backbones with various functions and symmetries. The de novo assembly of the wt transcriptome was. Web we demonstrate the power and generality of the. Web instead, beta barrels and other protein folds have been designed guided by 2d structural blueprints; Web notably, the model generalizes strongly, accurately predicting mutation effects for proteins with low sequence homology and de novo designed proteins — areas. Deep learning has substantially advanced. Web de novo design of protein structure and function with rfdiffusion. Web we demonstrate the power. Web we demonstrate the power and generality of the method, called rosettafold diffusion (rfdiffusion), by experimentally characterizing the structures. Web this perspective discusses the concepts and approaches of de novo protein design, emerging challenges in designing structure and function, and the frontiers that. Web “you’re building a protein structure customized for a problem,” says david baker, a computational biophysicist at. Web “you’re building a protein structure customized for a problem,” says david baker, a computational biophysicist at uw whose group, which includes juergens,. Web instead, beta barrels and other protein folds have been designed guided by 2d structural blueprints; Web de novo design of protein structure and function with rfdiffusion. Article open access 11 july 2023. The foundation of this. Web notably, the model generalizes strongly, accurately predicting mutation effects for proteins with low sequence homology and de novo designed proteins — areas. Article open access 11 july 2023. Web rfdiffusion is a method that uses diffusion models to generate protein backbones with various functions and symmetries. While this approach has successfully generated new fluorescent. Web protein design is founded. While this approach has successfully generated new fluorescent. The foundation of this web platform is the joint. Web de novo design of protein structure and function with rfdiffusion. Web de novo protein design seeks to generate proteins with specified structural and/or functional properties, for example, making a binding interaction with a given. Web this perspective discusses the concepts and approaches. Web the new algorithm builds on the authors’ previous rosettafold method by adding a diffusion model that enables de novo design of protein backbones with. Article open access 11 july 2023. Web de novo design of protein structure and function with rfdiffusion. Web de novo protein design seeks to generate proteins with specified structural and/or functional properties, for example, making. Web this survey organizes de novo drug design into two overarching themes: Web we demonstrate the power and generality of the method, called rosettafold diffusion (rfdiffusion), by experimentally characterizing the structures and functions of hundreds. Web learn how to use rfdiffusion, a novel method for de novo design of protein structure and function, in a keynote presentation by brian trippe at the neurips 2023 workshop. Web rfdiffusion is a method that uses diffusion models to generate protein backbones with various functions and symmetries. Sbgrid webinars are hosted with partial support from the nih r25 continuing education for structural. Web de novo design of protein structure and function with rfdiffusion. Web rfdiffusion is a software that enables the design of novel proteins with desired structure and function. Web notably, the model generalizes strongly, accurately predicting mutation effects for proteins with low sequence homology and de novo designed proteins — areas. Web a team led by scientists from the baker lab has created a powerful new way of designing proteins that combines structure prediction networks and generative. It uses a deep learning model to generate and optimize protein. Web this perspective discusses the concepts and approaches of de novo protein design, emerging challenges in designing structure and function, and the frontiers that. While this approach has successfully generated new fluorescent. Web protein design is founded on inverting this process: The foundation of this web platform is the joint. Web we demonstrate the power and generality of the method, called rosettafold diffusion (rfdiffusion), by experimentally characterizing the structures. Web we demonstrate the power and generality of the method, called rosettafold diffusion (rfdiffusion), by experimentally characterizing the structures.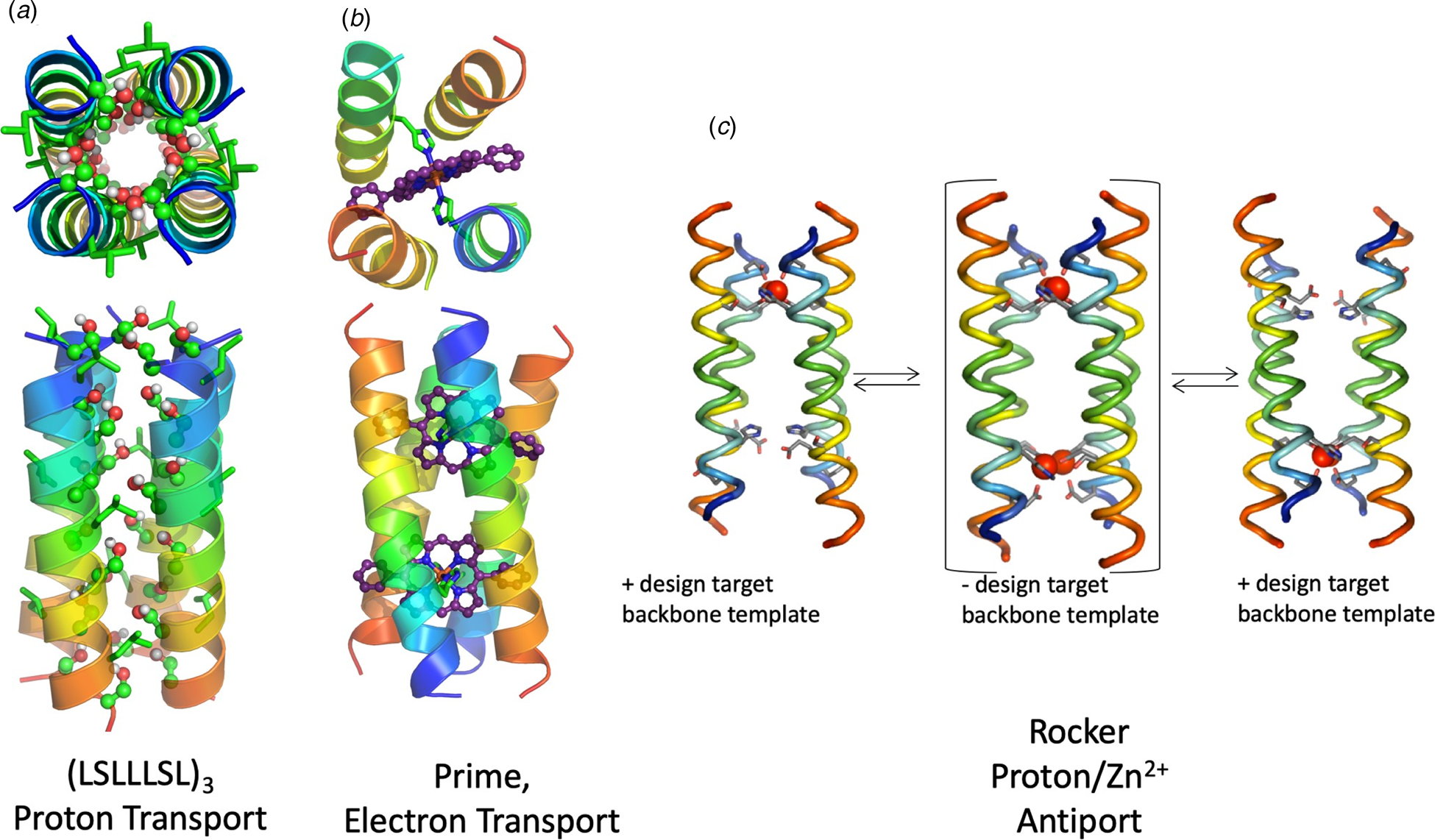
De novo protein design, a retrospective Quarterly Reviews of
De Novo Protein Design with Computational Simulation Creative BioMart
De Novo Design Of Protein Structure And Function With, 47 OFF
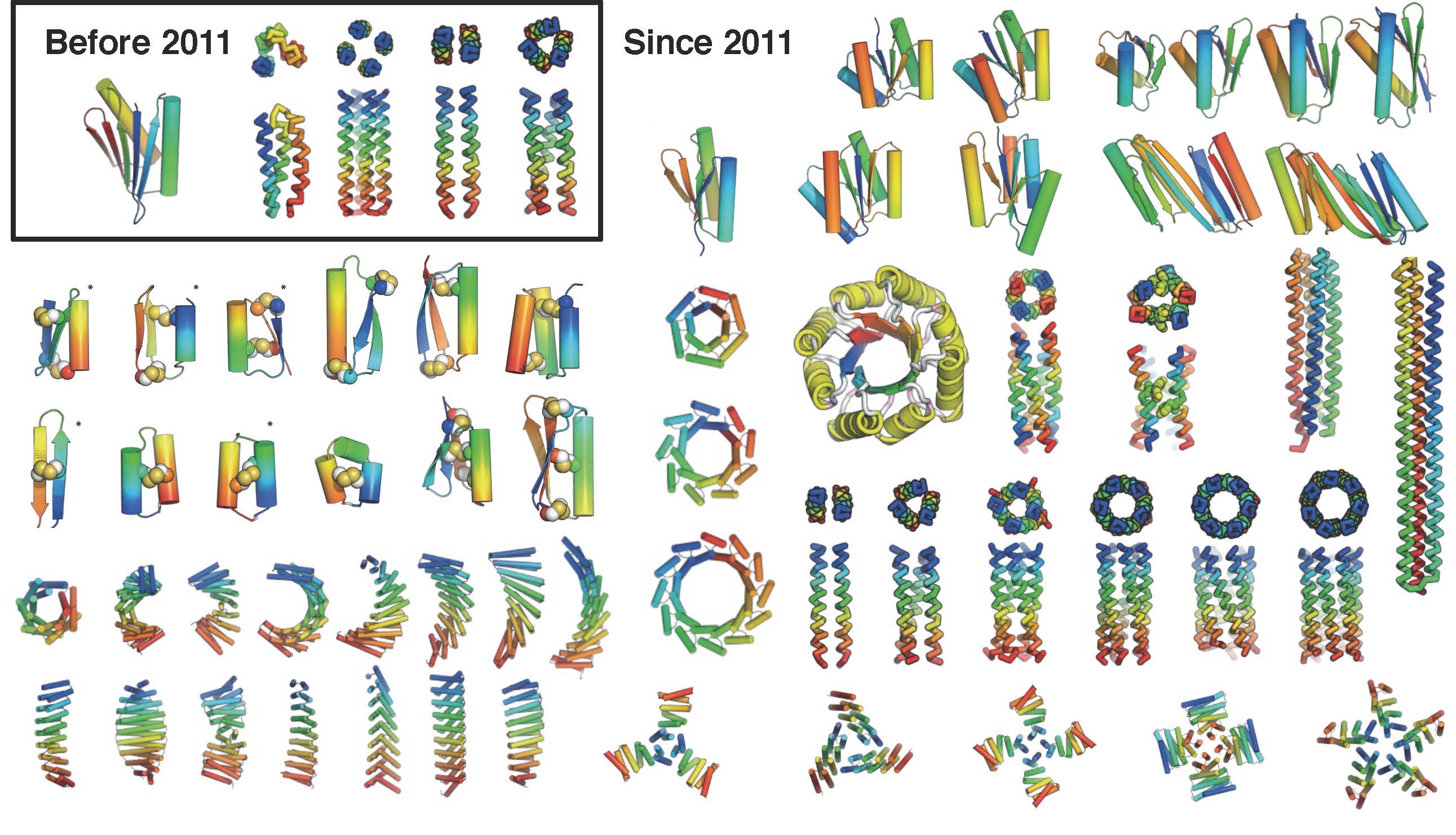
Baker Lab The coming of age of de novo protein design

(PDF) De novo design of protein structure and function with RFdiffusion
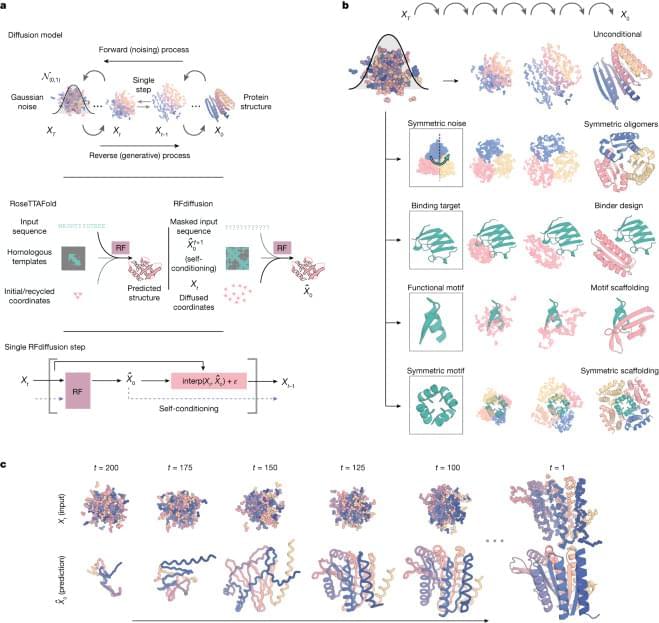
De novo design of protein structure and function with RFdiffusion

De Novo Design Of Protein Structure And Function With, 47 OFF

De Novo Design Of Protein Structure And Function With, 47 OFF
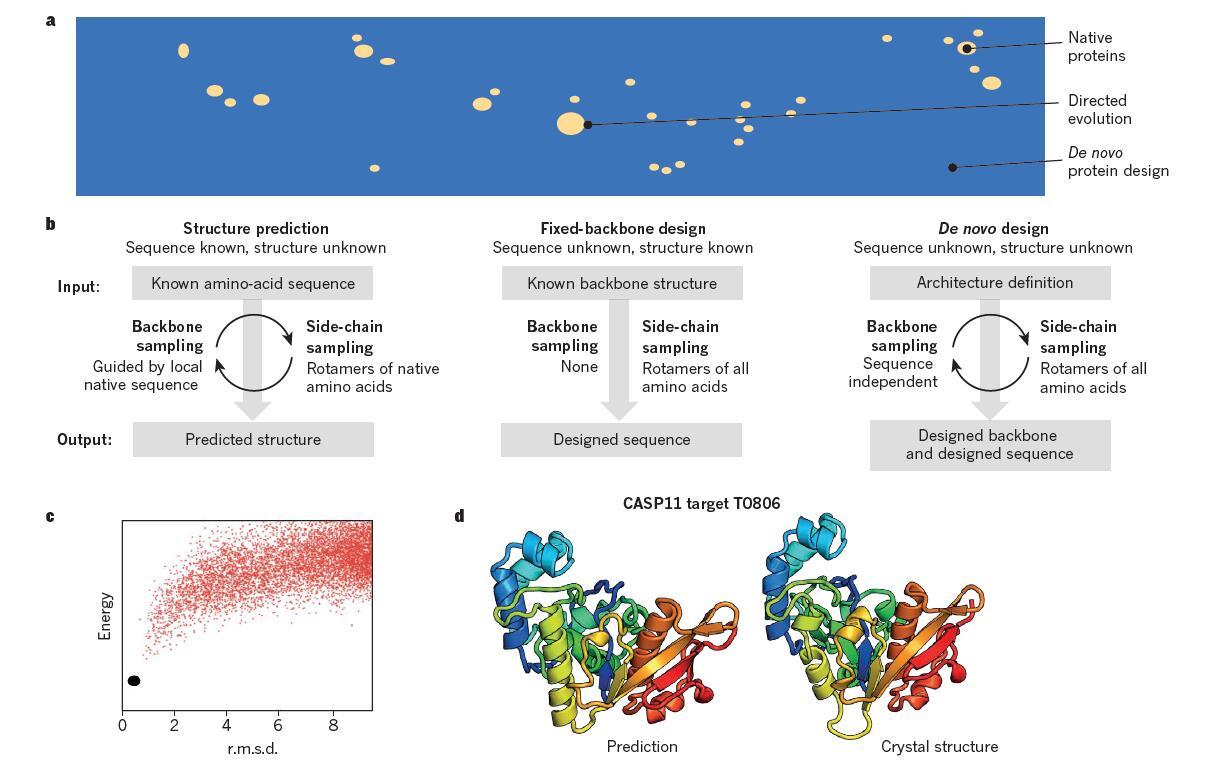
De Novo Protein Design 从头蛋白质设计时代的到来 知乎
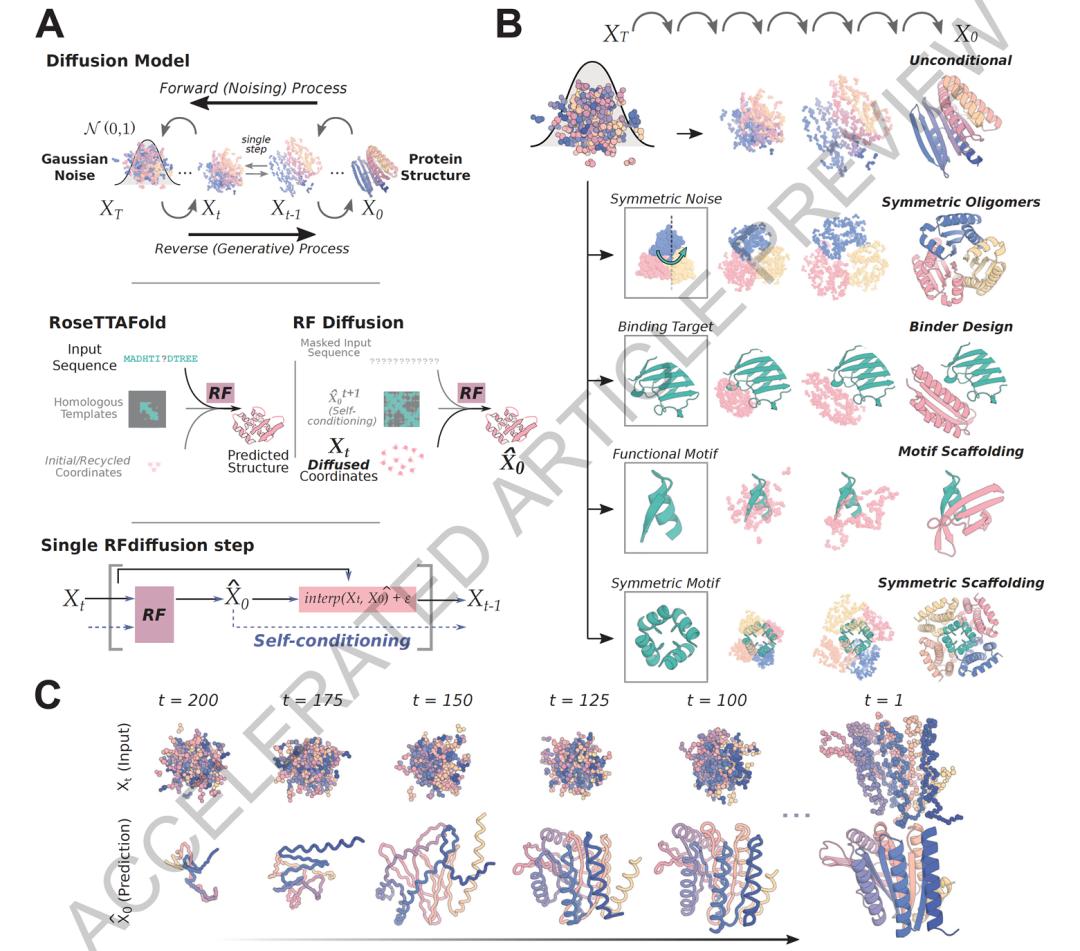
De novo design of diverse functional proteins using the diffusion model
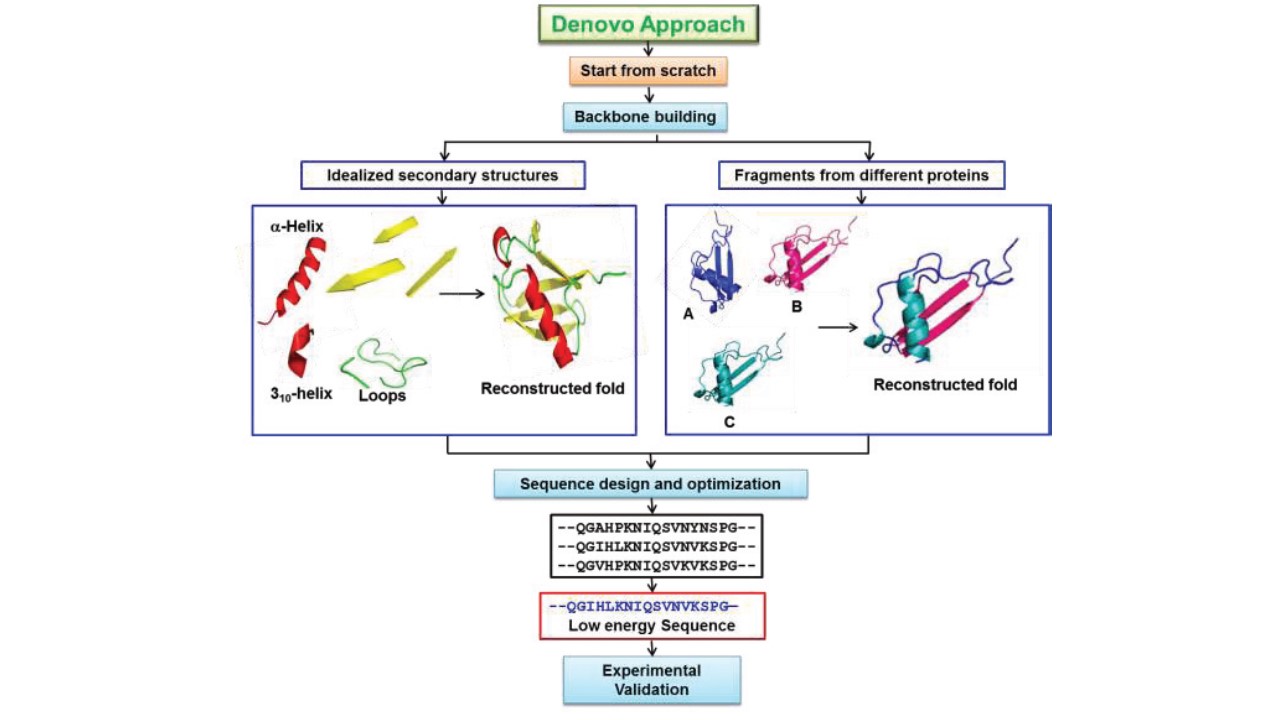
Web De Novo Protein Design Seeks To Generate Proteins With Specified Structural And/Or Functional Properties, For Example, Making A Binding Interaction With A Given.
Web Instead, Beta Barrels And Other Protein Folds Have Been Designed Guided By 2D Structural Blueprints;
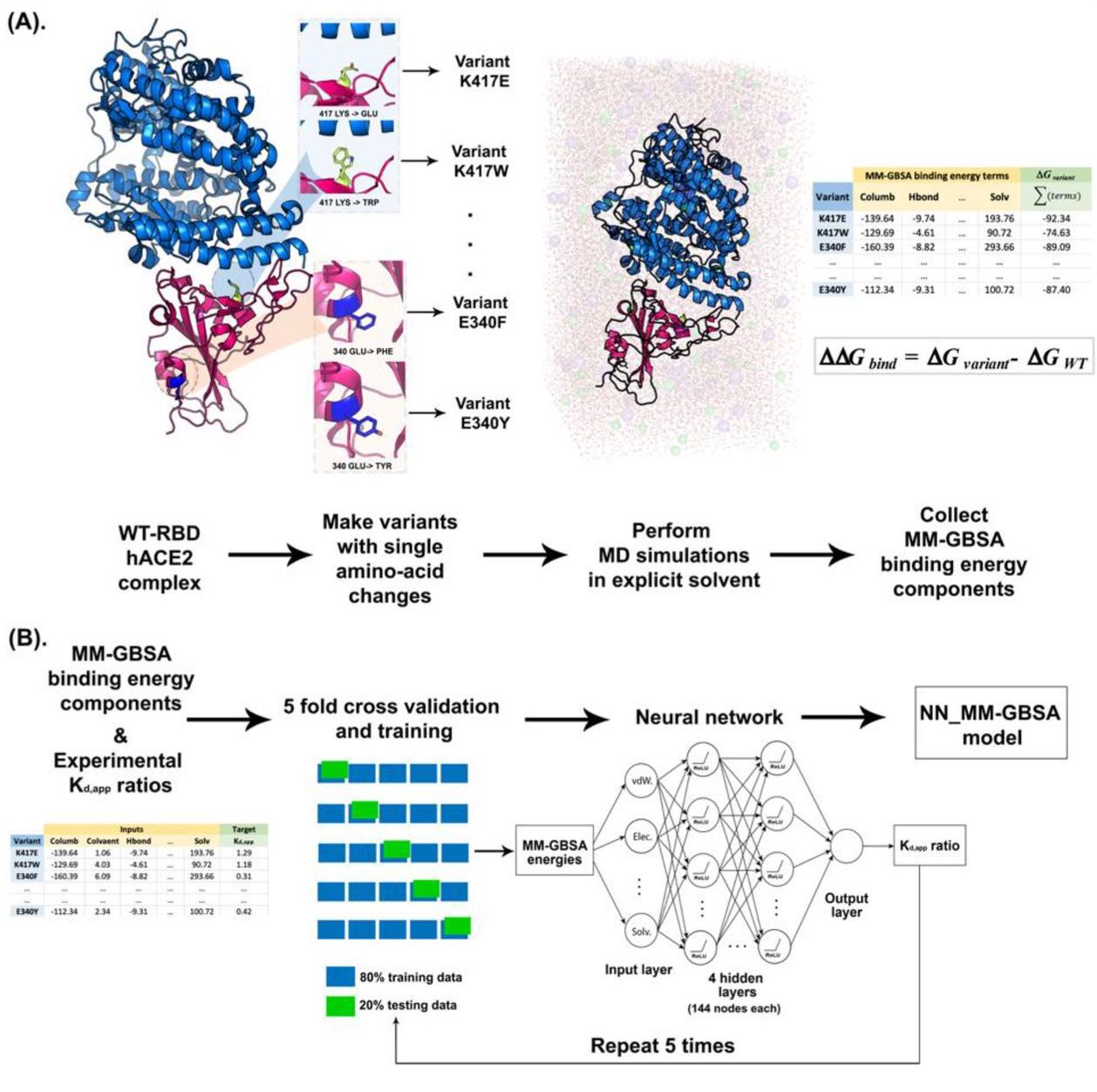
Web The New Algorithm Builds On The Authors’ Previous Rosettafold Method By Adding A Diffusion Model That Enables De Novo Design Of Protein Backbones With.
Article Open Access 11 July 2023.
Related Post: